1st Prize Winner: Felix Mikus, EMBL Heidelberg
 I obtained my Master’s degree in molecular and cellular biology from the University of Heidelberg. For my thesis project I worked with Freddy Frischknecht and Ross Douglas (now at University Giessen) on the fascinating cell biology of Plasmodium gametogenesis, which really helped shape my future research. In the autumn of 2020 I started my PhD at EMBL Heidelberg in the group of Gautam Dey, that studies the cell biology of the nucleus in an evolutionary context. Using the fission yeast Schizosaccharomyces pombe as the model organism of choice, my project focuses on the homeostasis of nuclear pore complexes (NPCs) and how their remodelling might regulate nuclear envelope breakdown. S. pombe divides its genome within an intact nuclear envelope, with breakdown only occurring in a narrowly defined zone, and we are investigating the hypothesis that this particular region is also utilised to remove old or damaged proteins in a process regulated by the NPC basket.
I obtained my Master’s degree in molecular and cellular biology from the University of Heidelberg. For my thesis project I worked with Freddy Frischknecht and Ross Douglas (now at University Giessen) on the fascinating cell biology of Plasmodium gametogenesis, which really helped shape my future research. In the autumn of 2020 I started my PhD at EMBL Heidelberg in the group of Gautam Dey, that studies the cell biology of the nucleus in an evolutionary context. Using the fission yeast Schizosaccharomyces pombe as the model organism of choice, my project focuses on the homeostasis of nuclear pore complexes (NPCs) and how their remodelling might regulate nuclear envelope breakdown. S. pombe divides its genome within an intact nuclear envelope, with breakdown only occurring in a narrowly defined zone, and we are investigating the hypothesis that this particular region is also utilised to remove old or damaged proteins in a process regulated by the NPC basket.
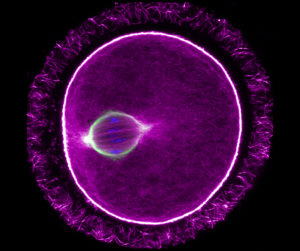
 This image is a time series projection of the acellular slime mould Physarum polycephalum (a.k.a. The Blob) colonising an agar plate in search of food. Powerful actomyosin contractions allow this single, giant cell – containing tens of thousands of nuclei sharing a single cytoplasm – to “expand” at several centimetres per hour, making it large enough to be photographed using cell phone cameras (in this case an iPhone 11). Our work with this organism, that has been largely forgotten by cell biologists for several decades, started as a short group sabbatical. For two weeks, each group member focused their work on particular aspects of its biology or on advancing methodology to kickstart future projects.
This image is a time series projection of the acellular slime mould Physarum polycephalum (a.k.a. The Blob) colonising an agar plate in search of food. Powerful actomyosin contractions allow this single, giant cell – containing tens of thousands of nuclei sharing a single cytoplasm – to “expand” at several centimetres per hour, making it large enough to be photographed using cell phone cameras (in this case an iPhone 11). Our work with this organism, that has been largely forgotten by cell biologists for several decades, started as a short group sabbatical. For two weeks, each group member focused their work on particular aspects of its biology or on advancing methodology to kickstart future projects.
2nd Prize Winner: Sam Dunkley, University of Bristol
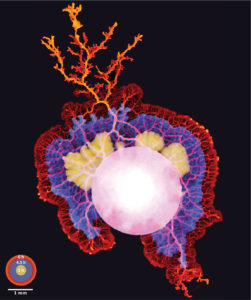
 I am a fourth year Dynamic Molecular Cell Biology PhD student at the University of Bristol. I completed my undergraduate BSc at the University of Bristol, before undertaking a master by research at the University of Oxford. Our lab works to unravel the molecular mechanisms at play throughout female meiosis and therefore understand the perturbations that leads to chromosome segregation abnormalities. We employ gain/loss of function assays in combination with high resolution live-imaging at different stages of meiosis. My research investigates the emerging role of the actin cytoskeleton in this fascinating process.
I am a fourth year Dynamic Molecular Cell Biology PhD student at the University of Bristol. I completed my undergraduate BSc at the University of Bristol, before undertaking a master by research at the University of Oxford. Our lab works to unravel the molecular mechanisms at play throughout female meiosis and therefore understand the perturbations that leads to chromosome segregation abnormalities. We employ gain/loss of function assays in combination with high resolution live-imaging at different stages of meiosis. My research investigates the emerging role of the actin cytoskeleton in this fascinating process.
Single slice Airyscan image of a mouse oocyte arrested at metaphase-I of Meiosis. Actin filaments can be seen permeating the microtubule-based spindle. Chromosome labelled with Hoechst (blue), the microtubule spindle by alpha-tubulin (green) and actin with phalloidin (magenta). The image was taken using a Zeiss LSM 800 with Airyscan.
3rd Prize Winner: Tom Mitchell, Queen Mary University London
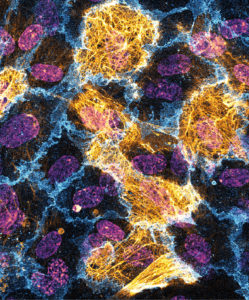
I studied Pharmacology as an undergraduate in King’s College London and had a particular interest in cardiovascular pharmacology. This led (after a period working in scientific publishing) to me applying for a cardiovascular-focused Masters’s and PhD programme at Queen Mary University of London. My PhD studies, in Tom Nightingale’s Lab, were focused on endothelial biology and I am continuing this project as a postdoc there. This research utilises a novel labelling technique to identify if changes in the trafficking of key proteins can lead to major alterations in endothelial cell function – most particularly in blood vessel growth.
Proximity labelling is a powerful tool to investigate protein trafficking. In these HUVECs a fusion protein of neuropilin-1 and HRP biotinylated proteins within a 20 nm radius (streptavidin, gold). This reveals a highly striated pattern of bounded by, and overlapping the cell junctions (PECAM-1, blue). Nuclei visualised via DAPI (Purple).